|
 What's ESPRESSO?
What's ESPRESSO?
Recombinant protein technology is an important issue in many industrial
and pharmacological applications. Nevertheless, the success rate of
obtaining soluble protein is relatively low, as many proteins do not
express or form inclusion body. In some cases, these problems are
overcome by optimizing the expression conditions, but a knowledge of
solubility under "standard" condition may provide a clue for
determining priority targets for experiments such as a large scale
proteomics analysis.
ESPRESSO (EStimation of PRotein ExpreSsion and SOlubility) predicts
protein expression and solubility from sequence information. ESPRESSO
makes two protein expression system, which are Escherichia Coli and
Wheat germ cell-free system, applicable to prediction. ESPRESSO
implicated two kinds of prediction methods. One is based on selected
property of sequence/structure, the other is based on short motifs
associated with protein expression or solubility. Refer to the Methods page for details.
 How to use ESPRESSO
How to use ESPRESSO
ESPRESSO provides a simple submission system.
At the beginning, click "SUBMISSION" tab, then you can submit your sequence in the form as below.
First, select three options, and then input query sequence.
Finally, you click "submit"
button.
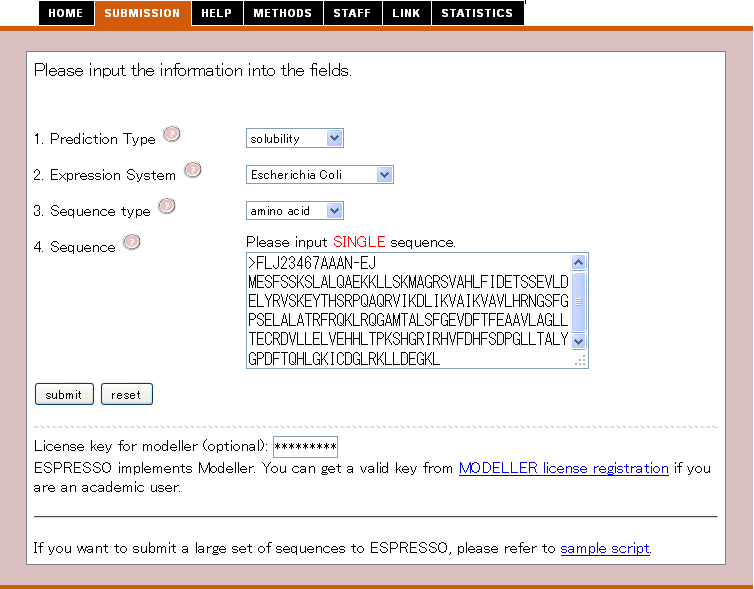
Image of submission page
|
1. Prediction Type
User selects the type of prediction
| expression |
It is predicted whether a sequence is expressed or not.
|
| solubility |
It is predicted whether a sequence is contained in soluble fraction or not.
|
2. Expression System
User selects the type of protein expression system.
| Escherichia Coli |
Escherichia Coli is used as a host cell.
|
| Wheat germ cell-free |
The wheat germ cell-free expression system is used for protein production.
|
| Brevibacillus |
The brevibacillus exepression system used as a host cell.
|
Refer to the Methods Page for the detailed information about experimental condition.
3. Sequence Type
User selects the type of sequence that you input into the sequence box below.
| nucleotide |
The sequence in the sequence box is considered to be nucleotide sequence.
|
| amino acid |
The sequence in the sequence box is considered to be amino acid sequence.
|
4. Sequence
Please input a nucleotide or an amino acid sequence to the sequence data box by a single sequence in plain text or in FASTA format.
ESPRESSO accepts the nucleotide sequence with from 60 nt to 3000 nt or amino acid sequence with from 20 aa to 1000 aa.
Prediction method changes with the combination of prediction type, expression system, and sequence type.
| | Escherichia Coli | Wheat germ | Brevibacillus |
| Expression | Solubility | Expression | Solubility | Expression | Solubility |
| Sequence type | Nucleotide | ○ | ○ | × | ○ | × | ○ |
| Amino acid | △ | ○ | × | ○ | × | ○ |
| ○ : |
Two prediction methods (property based prediction and motif based prediction) are executed.
|
| △ : |
Only motif basd prediction is preformed.
|
| × : |
No prediction is provided.
|
 Result Page
Result Page
The result page is consisted of three sections.
1. Prediction Summary
The first part shows the summary of protein expression or solubility prediction.
| Sequence |
The submitted sequence is displayed. When you submit a nucleotide sequence, the sequence translated into protein is displayed.
|
| Expression system |
Selected protein expression system.
|
| Property based prediction |
The prediction result is based on properties of sequence/structure. The
value in the parenthesis shows that reliability of prediction.
When you input amino acid sequence for predicting protein expression in
Escherichia Coli and sequence for predicting protein expression in wheat germ cell-free,
this mode cannot be executed.
|
| Motif based prediction |
The prediction result is based on the number of motifs associated with protein expression/solubility.
|
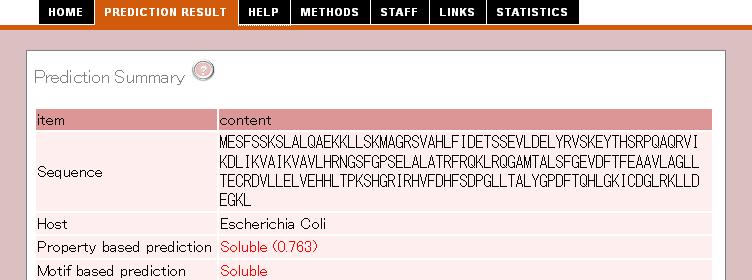
Image of Prediction Summary section
|
2. Picture View
The second part
illustrates the location of motifs associated with protein
expression/solubility, domains defined by Pfam-A, and predicted
secondary structure.
If you click "Go to Details" button, you can also see the same
information in sequence level.
| Motif |
The motif(s) associated with protein expression/solubility are displayed.
The red bar(s) show the positive effect of protein expression/solubility.
Inversely, the blue bar(s) show the negative effect.
|
| Pfam |
The conserved domain(s) are displayed.
These domains are detected by searching Pfam-A1 database with hmmscan2.
A longer domain is chosen by priority.
|
| SS(psipred) |
The secondary structure signifies by using PHD3.
The green and yellow bar(s) are corresponding to alpha-helix and beta-sheet, respectively.
|
| Disorder (POODLE-L) |
The putative disordered region(s) predicted by POODLE-L4 are displayed ub tge cyan bar(s).
|
| PDB (BLAST) |
The structured region(s) determined by X-ray or NMR are displayed. These are detected bu searching the pdbaa with BLAST. A longer structure is chosen bu priority.
|
1 Finn RD. et al. Nucleic Acids Res. 2008 Jan 1;36(Database issue):D281-8.
2 Deey SR. PLoS Comput. Biol. 2008 May 30;4(5):e1000069.
3 Rost B. Method Enzymol. 1996;266:525-39.
4 Hirose S. et al. Bioinformatics 2007 Aug 15;23(16).
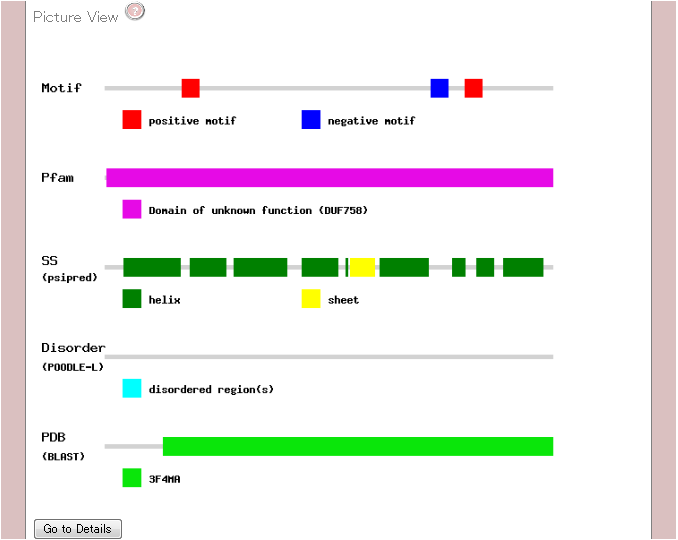
Image of Picture View section
|
The location of motif and other structural information are useful for a user to modify sequence.
3. Structure View
The third part illustrates the location of motifs associated with protein expression/solubility in the structure.
If you clike the "PyMol file" button, you can download the pymol file.
If a similar sequeunce in PDB are not found, there is no data in the structure view.
| Template |
PDB id which is used as the template of homology modeling.
|
| Target region |
The region where the modeling structure be created.
|
| Template region |
The region used for the homology modeling.
|
| Sequence id |
The sequence identity between query sequence and template structure.
|
| E-value |
The E-value score when BLAST search executes against pdbaa.
|
| Model score (MODELLER) |
The score obtained br MODELLER. The hihger score indicates the higher accurate modeling structure.
|
| structure view |
The motif(s) associated with protein expression/solubility are displayed in the strucutre.
The red shows the positive effect of protein expression/solubility.
Conversely, the blue shows the negative effect.
|
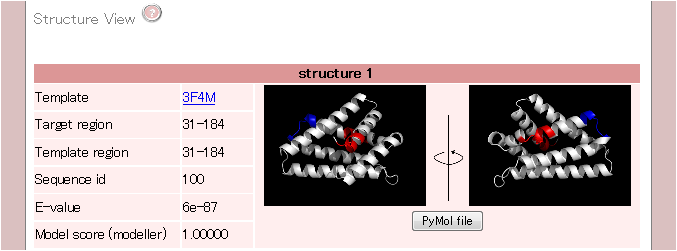
Image of Structure View section
|
If you do not enter the modeller licence key in the submission page, you can not see the result.
Please get MODELLER licence if you are an academic user.
4. Supplementary Data
The fourth part lists the location and sequence of motifs associated with protein expression/solubility.
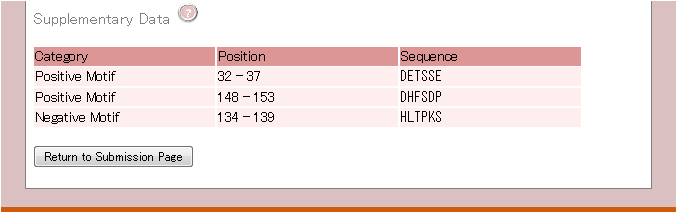
Image of Supplementary Data section
|
5. The proposals for increasing/decreasing properties of expression or solubility (New function)
The fifth part lists nucleotides or amino acids, which increase/decrease properties of expression or solubility, are proposed in order of position.
If you click the proposed nucleotide or amino acid, ESPRESSO will start to predict with the mutated sequence. In the result of ESPRESSO, the nucleotides or the amino acids are proposed again. You can increase/decrease properties of expression or solubility more and more by repeat operation.
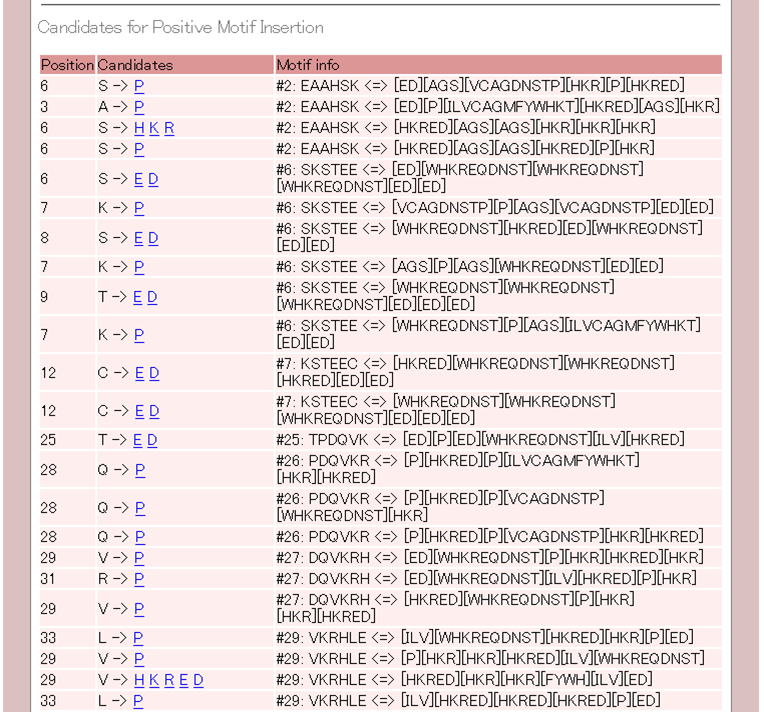
The amino acids to increase the solubility
|
Please refresh the "Under Calculation page" manually, when you cannot
obtain prediction result even if you wait for a long time. In Safari and Google Chrome, this error sometimes occures. We recommend that users use Internet Explorer or FireFox.
 Template Script for Accessing ESPRESSO
Template Script for Accessing ESPRESSO
The template script can access and perform the service of ESPRESSO.
The script is written in Perl.
The script requires an input file and creates two kinds of output files.
Requirement
The template script utilized two perl modules:
LWP (libwww-perl) and
URI .
They are downloaded from CPAN (http://www.cpan.org/).
Please install them into your PC.
Download Files
How to use the script
The first argument should be an input file.
$ perl sample.pl Input.txt
FLJ13012AAAN-EJ_AA
FLJ13012AAAN-EJ_nt
FLJ80169AAAN-EJ_AA
FLJ80169AAAN-EJ_AA_E
FLJ32966
FLJ32966_E
|
Input file
The input file includes information about query sequence and parameters.
| first column |
The sequence name or ID.
|
| second column |
The type of query sequence.
You must input "nt" for nucleotide sequence,
"AA" for amino acid sequence.
|
| third column |
The query sequence.
|
| fourth column |
The prediction type.
You must input "EXP" for protein expression prediction,
"SOL" for protein solubility prediction.
|
| fifth column |
The expression system.
You must input "Ecoli" for Escherichia Coli ,
"Wheat" for wheat germ cell-free, and "Brevi" for Brevibacillus expression system.
|
Output file
Output file 1 (the extension is ".ESPRESSO") includes summary of prediction results.
| first column |
The sequence name or ID.
|
| second column |
The query sequence.
|
| third column |
The expression system.
|
| fourth column |
The prediction result of property based prediction.
|
| fifth column |
The probability of property based prediction.
|
| sixth column |
The prediction result of motif based prediction.
|
| seventh column |
The motifs that have positive influence on protein expression or solubility, and their position on sequence.
|
| eighth column |
The motifs that have negative influence on protein expression or solubility, and their position on sequence.
|
| ninth column |
The name of conserved domain and their position on sequence.
|
| tenth column |
The predicted secondary structure. H, E, and C respectively signify helix, sheet and coil.
|
Output file 2 (the extension is ".error") includes sequence ID and error code.
If an error has occurred, please refer to the following table.
| Input error |
There are some errors in the input file.
The prediction type or expression system is wrong.
The query sequence has wrong characters or cannot translate into amino acid.
Also, its length is very short or long.
|
| No prediction |
When you input amino acid sequence for predicting protein expression in Escherichia Coli
and sequence for predicting protein expression in wheat germ cell-free system,
the prediction cannot be performed.
|
| Calculation failed |
There were some troubles in ESPRESSO server. Please try again.
|
 Contact Us
Contact Us
Any comments or questions about ESPRESSO services send to noguchit(at)my-pharm(dot)ac(dot)jp.
|
|