|
. Prediction method . Prediction accuracy . How to use POODLE . Ackonwledgment . Publication . Contact Us What's POODLE-I ? POODLE series, which are POODLE-W, -L, and -W, are disordered region predictors depending on their length. Therefore, we have to consider of which prediction result is adopted each time. We assumed that the factor causing a short disordered region might be different from the factor causing a long ones: a short disordered region is mainly determined according to whether it is located within a structure such as a loop or linker. By contrast, a long disordered region is mainly affected to the physico-chemical property derived from the primary sequence such as low hydrophobicity or high charge content. According to the idea, we employed a workflow approach for combining prediction results from the POODLE series. Additionally, POODLE-I uses predicted structural information such as the secondary structure and similar structure on predicting disordered region.  Prediction Method POODLE-I employed workflow approach on predicting disordered region. An important point of the prediction method is that short and long disordered regions are detected separately, so that the flow chart is divisible into two parts. One side flow predicts a long disordered region using amino acid property, the other side predicts a short disordered regions using structural information. The flow chart requires information of amino acid sequence as input data, and assigns an order/disordered state to each residue. A similar structure is enclosed with a dashed line obtained from TS pred, which signifies the fold recognition method. COILpred, SSpred and ASApred, which are enclosed in thick kines respectively signify coiled coil predictor, secondary structure predictors, and accessible surface area predictors. The alphabet letters enclosed by a diamond represent branch on conditions. Respective conditions answer the following: (a) Is a query unfolded? (b) Does a query include long disordered region? (c) Is there any secondary structure in the terminal region of disordered region? (d) Is there any a coiled coil region in disordered region? (e) Does an ordered region predicted by POODLE-L and -W include a disordered region? (f) Can a similar structure by found? (g) Is there any missing region? and (h) Is there any secondary structure or buried region in disordered region? 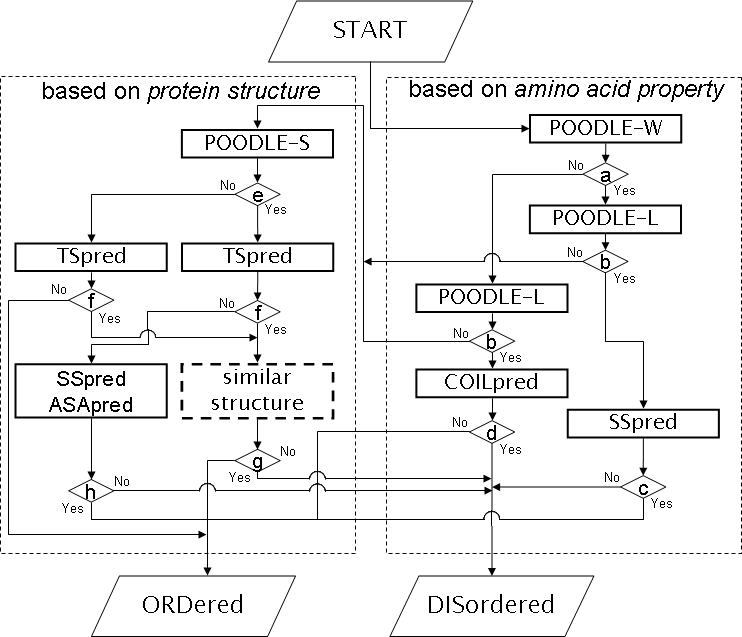 The programs implementing in POODLE-I are as follows.
 Prediction Accuracy Prediction accuracy was estimated and compared POODLE-I with POODLE series using 123 protein targets used in CASP8, representing 23,924 ordered residues and 2,926 disordered residues. The assessment was performed on a residue basis. Five criteria was adopted: sensitivity (= TP / (TP+FN)), specificity (=TP / (TP+FP)), accuracy (ACC) (= (sensitivity+specificity) / 2)), CASP weighted score (Sw) (= ((WdisTP-WordFP+WordTN-WdisFN) / (Wdis(TP+FP)+Word(TN+FN)), and the area under the ROC curve (AUC). Furthermore, TP, FP, FN, and TN respectively denote the numbers of true positive, false positive, false negative, and true negative results. In fact, Wdis and Word are weights which set to the rate of disordered and ordered residues in a dataset.
 How to use POODLE-I? 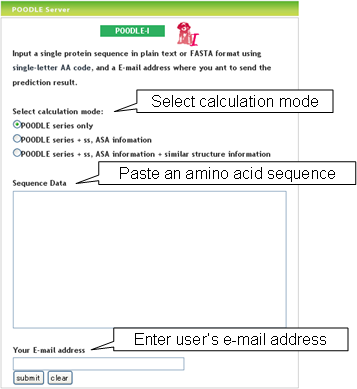 POODLE-I has three-step for prediction. In the first step, the user can
select which information is used for prediction. In the second step, the
user paste an amino acid sequence in FASTA format in the Sequence Data
box. The POODLE-I server receives only 20 characters representing
standard amino acids. In the third step, the user enters an accessible
e-mail address to which the prediction result is sent.
POODLE-I has three-step for prediction. In the first step, the user can
select which information is used for prediction. In the second step, the
user paste an amino acid sequence in FASTA format in the Sequence Data
box. The POODLE-I server receives only 20 characters representing
standard amino acids. In the third step, the user enters an accessible
e-mail address to which the prediction result is sent. POODLE-I server provides two types of output style. One is a plane text based on CASP format, which is included in a message body in E-mail. The other is a graphical output which is accessible from a link included in E-mail. The interface is the same as POODLE-L. Detailed information is provided from a POODLE-L help page.  Acknowledgment This work was supported by PharmaDesign, Inc.  Publication S.Hirose, K.Shimizu, N.Inoue, S.Kanai and T,Noguchi "Disordered region prediction by integrating POODLE series", CASP8 Proceedings 2008, 14-15.  Contact Us   |